The first successful biological synthesis of antibiotic analogs opens new avenues in the fight against tuberculosis.
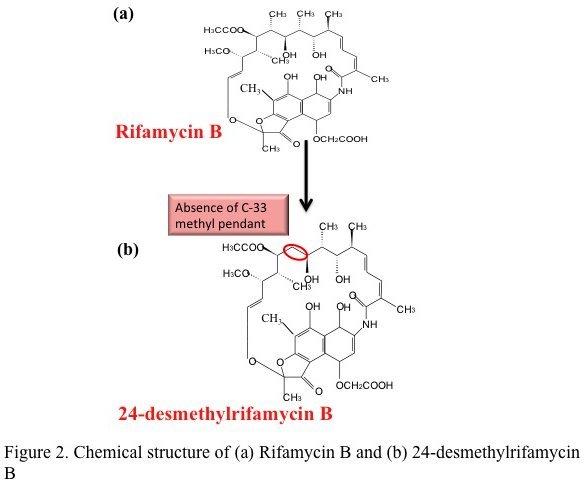
A ray of hope against tuberculosis — a multi-lab team lead by Rup Lal at Delhi University has reported the first successful bacterial synthesis of a rifamycin analog. This study published in the Journal of Biological Chemistry describes the genetic modification of the natural producer of rifamycin — the bacterium Amycolatopsis mediterranei to produce the analog 24-desmethyl rifamycin B.
Rifampicin is a semisynthetic derivative of rifamycin B, which since its discovery in 1960s has been the mainstay for TB treatment. Rifampicin inhibits prokaryotic RNA polymerase and thereby DNA dependent RNA synthesis — a key feature that makes the antibiotic effective against even latent Mycobacterium. Many derivatives of rifamycin B have been synthesized over the past decades, some effective, some less so. As the pathogens eventually develop resistance to each new analog, there is perpetual demand for new antibiotics and analogs.
“Chemists couldn’t do much”, said Prof. Lal. “Rifamycin B has a complex aromatic ring structure, with a naphthalene core spanned by an aliphatic chain. This structure makes it less conducive to chemical modifications,” he added.
The sequential biosynthesis of the antibiotic in bacteria provided a gateway. The antibiotic is synthesized by an operon of 5 genes called type 1 polyketide synthases (PKS). These genes add acetate or propionate units to an aromatic core structure to synthesize rifamycin. According to Prof. Lal, “It is possible to replace the domain that adds acetate with that of the one that adds propionate, thereby tricking the bacteria to produce an analog”.
In theory, this seemed simple — genes can be swapped by homologous recombination, commonly done in other bacteria. A. mediterranei, however proved notoriously difficult to manipulate. It took the team nearly 20 years to generate a clone of the bacteria producing the desired analog. This clone had replaced the indigenous gene that adds propionate to the growing chain with a gene from the rapamycin PKS operon that adds acetate. Chemically the mutant produced 24-desmethyl rifamycin B (simplistically rifamycin minus a methyl group). This was then chemically converted to two of its semisynthetic derivatives i.e. 24-desmethyl rifamycin S and 24-desmethyl rifampicin.
The analogs of rifamycin B and its derivatives analyzed by mass spectrometry showed the expected difference in the masses compared to rifamycin B. The structure of the analogs was further confirmed by NMR studies done in collaboration with Taifo Muhmud of Oregon State University.
When the analogs were assayed for their antibacterial activity, both 24-desmethylrifamycin S and 24-desmethylrifampicin proved effective different bacteria, including Staphylococcus. More satisfyingly, the MDR strain of Mycobacterium tuberculosis was also susceptible.
Though exciting, it is still early stages. While the team prepares for mice and clinical studies, it is also working towards increasing the yields of the analog. Currently, the yield is nearly 1000X lower than commercial production of rifamycin from A. mediterranei. This could be a huge drawback if the analogs were to be produced as a therapeutic drug. Lal, however, is very optimistic. He feels that yields can be increased, simply by producing the analog in the industrial strain of the bacteria. This however needs to be tested.
Regardless of how and when this analog hits the market as a treatment option, this study has surely broken the shackles of cumbersome chemical modifications and shown the possibility of synthesizing antibiotic analogs biologically, thereby opening the doors to development of myriad modifications and analogs to continue the fight against tuberculosis.